Continuum Removal
Anonym
Recently, I was given the chance to practice some
spectroscopy and in preparation for the project, I realized that I did not have
a simple way to visualize the variations in different absorption features
between very discreet wavelengths. The method that I elected to employ for this
task is called continuum removal
(Kokaly, Despain, Clark, & Livo, 2007). This method allows you to
compare different spectra and essentially normalize the data so that they can
be more easily compared with one another.
To use the algorithm, you first select the region that you
are interested in (for me this was between 550 nm and 700 nm -this is the
region of my spectra that deals with chlorophyll absorption and pigment). Once
the region is selected then a linear model is fit between the two points and
this is called the continuum. The continuum is the hypothetical background
absorption feature, or artifact or other absorption feature, which acts as the
baseline for the target features to be compared against (Clark, 1999). Once the
continuum has been set then the continuum removal process can be performed on
all spectra in question using
the following equation (Kokaly,
Despain, Clark, & Livo, 2007).
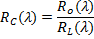
RC is
the resulting continuum removed spectra, RL is the continuum line
and, Ro is the original reflectance value.
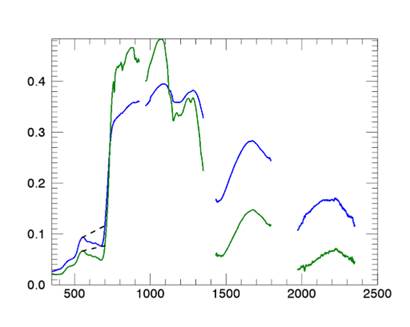
Figure 1:
Original spectra of two healthy plants. The dotted line denotes the continuum
line. The x axis shows wavelengths in nm and the y axis represents reflectance.
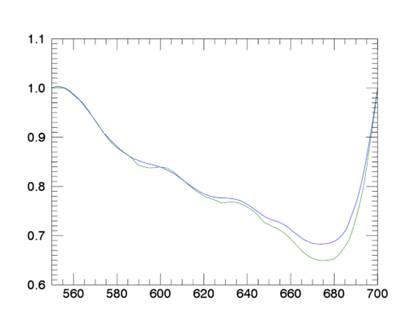
Figure 2: The
continuum removal for wavelengths 550 nm - 700 nm.
The resulting code gives you a tool that will take in two
spectral libraries, with one spectra per library, and return two plots similar
to what is shown in Figure 1 and Figure 2.
pro Continuum_Removal
compile_opt IDL2
Spectra_File_1 =
Spectra_File_2 =
;
Find the bounds for the feature
FB_left = 550
FB_right =700
;
Open Spectra 1
oSLI1 = ENVISpectralLibrary(Spectra_File_1)
spectra_name =
oSLI1.SPECTRA_NAMES
Spectra_Info_1 =
oSLI1.GetSpectrum(spectra_name)
;
Open Spectra 2
oSLI2 = ENVISpectralLibrary(Spectra_File_2)
spectra_name =
oSLI2.SPECTRA_NAMES
Spectra_Info_2 =
oSLI2.GetSpectrum(spectra_name)
; Get
the wavelengths
wl =
Spectra_Info_1.wavelengths
;
Create Bad Bands List (this removes some regions of the spectra associated with
water vapor absorption)
bb_range = [[926,970],[1350,1432],[1796,1972],[2349,2500]]
bbl = fltarr(n_elements(wl))+1
dims = size(bb_range, /DIMENSIONS)
for i = 0
, dims[1]-1
do begin
range =
bb_range[*,i]
p1 = where(wl eq
range[0])
p2 = where(wl eq
range[1])
bbl[p1:p2] = !VALUES.F_Nan
endfor
;Plot
oSLI1 / oSLI2
p = plot(wl, Spectra_Info_1.spectrum*bbl,
xrange = [min(wl, /nan),max(wl, /nan)],$
yrange=[0,max([Spectra_Info_1.spectrum*bbl,Spectra_Info_2.spectrum*bbl],
/nan)], thick = 2, color = 'blue')
p = plot(wl, Spectra_Info_2.spectrum*bbl,
/overplot, thick = 2, color = 'green')
;
create the linear segment
Spectra_1_y1 =
Spectra_Info_1.spectrum[where( wl
eq FB_left)]
Spectra_1_y2 =
Spectra_Info_1.spectrum[where( wl
eq FB_right)]
pl_1 = POLYLINE([FB_left,FB_right], [Spectra_1_y1,
Spectra_1_y2], /overplot, /data, thick = 2,
LINESTYLE = '--')
Spectra_2_y1 =
Spectra_Info_2.spectrum[where( wl
eq FB_left)]
Spectra_2_y2 =
Spectra_Info_2.spectrum[where( wl
eq FB_right)]
pl_2 = POLYLINE([FB_left,FB_right], [Spectra_2_y1,
Spectra_2_y2], /overplot, /data, thick = 2,
LINESTYLE = '--')
; Get
the equation of the line
LF_1 = LINFIT([FB_left,FB_right], [Spectra_1_y1,
Spectra_1_y2])
LF_2 = LINFIT([FB_left,FB_right], [Spectra_2_y1,
Spectra_2_y2])
; Get
the values between the lower and upper bounds
x_vals = wl [ where(wl eq
FB_left) : where(wl eq FB_right)]
;
Compute the continuum line
RL_1 = LF_1[0] + LF_1[1]*
x_vals
RL_2 = LF_2[0] + LF_2[1]*
x_vals
;
Perform Continuum Removal
Ro_1 =
Spectra_Info_1.spectrum[ where(wl
eq FB_left) : where(wl eq
FB_right)]
RC_1 = Ro_1 / RL_1
Ro_2 =
Spectra_Info_2.spectrum[ where(wl
eq FB_left) : where(wl eq
FB_right)]
RC_2 = Ro_2 / RL_2
;
Plot the new Continuum Removal Spectra
pl_RC_1 = plot(x_vals, RC_1, color = 'Blue', xrange = [min(x_vals,
/NAN), max(x_vals, /NAN)] )
pl_RC_2 = plot(x_vals, RC_2, color = 'Green', /overplot)
end
Kokaly, R. F., Despain, D. G., Clark, R. N., & Livo, K.
E. (2007). Spectral analysis of absorption features for mapping vegetation
cover and microbial communities in Yellowstone National Park using AVIRIS data.
Clark, R. N. (1999). Spectroscopy of rocks and minerals, and
principles of spectroscopy. Manual of remote sensing, 3,
3-58.