Smoothing is used to reduce noise or to produce a less pixelated image. Most smoothing methods are based on low-pass filters, but you can also smooth an image using an average or median value of a group of pixels (a kernel) that moves through the image.
The following example uses the SMOOTH and MEDIAN functions with a moving average on a photomicrograph image of human red blood cells. This example data is available in the examples/data directory of your IDL installation.
The code shown below creates the following images, each displayed in separate windows.


Copy the entire code block and paste it into the IDL command line to run it.
file = FILEPATH('rbcells.jpg', $
SUBDIRECTORY = ['examples', 'data'])
READ_JPEG, file, rbcimage
img01 = IMAGE(rbcimage, LOCATION = [0, 0], $
TITLE = 'Red blood cells original image')
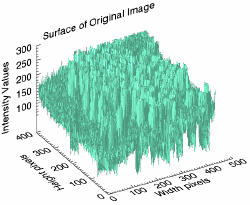
s1 = SURFACE(rbcimage, LOCATION = [500, 0], $
XTITLE = 'Width pixels', $
YTITLE = 'Height pixels', $
ZTITLE = 'Intensity Values', $
TITLE = 'Surface of Original Image', $
COLOR = 'aquamarine', $
ZTICKVALUES = [100, 150, 200, 250, 300])
(s1['zaxis']).location = [0, (s1.yrange)[1], 0]
smoothed_image = SMOOTH(rbcimage, 5, /EDGE_TRUNCATE)
img02 = IMAGE(smoothed_image, LOCATION = [0, 50], $
TITLE = 'Average-smoothed image')
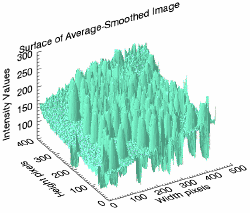
s2 = SURFACE(smoothed_image, location = [500, 50], $
XTITLE = 'Width pixels', $
YTITLE = 'Height pixels', $
ZTITLE = 'Intensity Values', $
TITLE = 'Surface of Average-Smoothed Image', $
COLOR = 'aquamarine', $
ZTICKVALUES = [100, 150, 200, 250, 300])
(s2['zaxis']).location = [0, (s2.yrange)[1], 0]
median_image = MEDIAN(rbcimage, 5)
img03 = image(median_image, LOCATION = [0, 100], $
TITLE = 'Median-smoothed image')
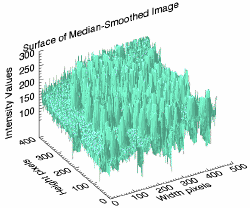
s3 = SURFACE(median_image, LOCATION = [500, 100], $
XTITLE = 'Width pixels', $
YTITLE = 'Height pixels', $
ZTITLE = 'Intensity Values', $
TITLE = 'Surface of Median-Smoothed Image', $
COLOR = 'aquamarine', $
ZTICKVALUES = [100, 150, 200, 250, 300])
(s3['zaxis']).location = [0, (s3.yrange)[1], 0]
Resources