The IMSL_ALLBEST procedure selects the best multiple linear regression models.
This routine requires an IDL Advanced Math and Stats license. For more information, contact your sales or technical support representative.
The IMSL_ALLBEST procedure finds the best subset regressions for a regression problem with
n_candidate = (N_ELEMENTS (x (0, *)))
independent variables. Typically, the intercept is forced into all models and is not a candidate variable. In this case, a sum-of-squares and crossproducts matrix for the independent and dependent variables corrected for the mean is computed internally. There may be cases when it is convenient for you to calculate the matrix; see the description of the COV_INPUT optional keyword.
"Best" is defined, on option, by one of the following three criteria:
-
R2 (in percent):
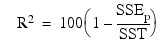
-
R2a (adjusted R2 in percent):
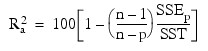
Note that maximizing the criterion is equivalent to minimizing the residual mean square:
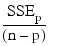
-
Mallows' Cp statistic:
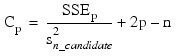
Here, n is equal to the sum of the frequencies (or N_ELEMENTS(x (*, 0)) if FREQUENCIES is not specified) and SST is the total sum of squares. SSEp is the error sum of squares in a model containing p regression parameters including β0 (or p – 1 of the n_candidate candidate variables). Variable is the s2n_candidate error mean square from the model with all n_candidate variables in the model. Hocking (1972) and Draper and Smith (1981, pp. 296–302) discuss these criteria.
The IMSL_ALLBEST procedure is based on the algorithm of Furnival and Wilson (1974). This algorithm finds MAX_N_BEST candidate regressions for each possible subset size. These regressions are used to identify a set of best regressions. In large problems, many regressions are not computed. They may be rejected without computation based on results for other subsets; this yields an efficient technique for considering all possible regressions.
There are cases when you may wish to input the variance-covariance matrix rather than allow the IMSL_ALLBEST procedure to calculate it. This can be accomplished using keyword COV_INPUT. Three situations in which you may want to do this are as follows:
- The intercept is not in the model. A raw (uncorrected) sum-of-squares and crossproducts matrix for the independent and dependent variables is required. Keyword COV_NOBS must be set to 1 greater than the number of observations. Form ATA, where A = [A, Y], to compute the raw sum-of-squares and crossproducts matrix.
- An intercept is to be a candidate variable. A raw (uncorrected) sum-of-squares and crossproducts matrix for the constant regressor (= 1.0), independent variables, and dependent variables is required for COV_INPUT. In this case, COV_INPUT contains one additional row and column corresponding to the constant regressor. This row/column contains the sum of squares and crossproducts of the constant regressor with the independent and dependent variables. The remaining elements in COV_INPUT are the same as in the previous case. Keyword COV_NOBS must be set to 1 greater than the number of observations.
- There are m variables to be forced into the models. A sum-of-squares and crossproducts matrix adjusted for the m variables is required (calculated by regressing the candidate variables on the variables to be forced into the model). Keyword COV_NOBS must be set to m less than the number of observations.
The IMSL_ALLBEST procedure saves considerable CPU time over explicitly computing all possible regressions. However, the procedure has some limitations that can cause unexpected results for users who are unaware of the limitations of the software.
- For n_candidate + 1 > –log2(e), where e is machine precision, some results may be incorrect. This limitation arises because the possible models indicated (the model numbers 1, 2, ..., 2n_candidate) are stored as floating-point values; for sufficiently large n_candidate, the model numbers cannot be stored exactly. On many computers, this means IMSL_ALLBEST (for n_candidate > 24; single precision) and IMSL_ALLBEST (for n_candidate > 49; double precision) can produce incorrect results.
- The IMSL_ALLBEST procedure eliminates some subsets of candidate variables by obtaining lower bounds on the error sum of squares from fitting larger models. First, the full model containing all n_candidate is fit sequentially using a forward stepwise procedure in which one variable enters the model at a time, and criterion values and model numbers for all the candidate variables that can enter at each step are stored. If linearly dependent variables are removed from the full model, error STAT_VARIABLES_DELETED is issued. If this error is issued, some submodels that contain variables removed from the full model because of linear dependency can be overlooked if they have not already been identified during the initial forward stepwise procedure. If error STAT_VARIABLES_DELETED is issued and you want the variables that were removed from the full model to be considered in smaller models, rerun the program with a set of linearly independent variables.
Example
This example uses a data set from Draper and Smith (1981, pp. 629-630). The IMSL_ALLBEST procedure is used to find the best regression for each subset size using the Mallow's Cp statistic as the criterion. Note that when Mallow's Cp statistic (or adjusted R2) is specified, the variable MAX_N_BEST indicates the total number of "best" regressions (rather than indicating the number of best regressions per subset size, as in the case of the R2 criterion). In this example, the three best regressions are found to be (1, 2), (1, 2, 4), and (1, 2, 3).
PRO IMSL_ALLBEST_ex1
x = transpose( [ [7., 26., 6., 60.], [1., 29., 15., 52.], $
[11., 56., 8., 20.], [11., 31., 8., 47.], $
[7., 52., 6., 33.], [11., 55., 9., 22.], $
[3., 71., 17., 6.], [1., 31., 22., 44.], $
[2., 54., 18., 22.], [21., 47., 4., 26.], $
[1., 40., 23., 34.], [11., 66., 9., 12.], $
[10., 68., 8., 12.]])
y = [78.5, 74.3, 104.3, 87.6, 95.9, 109.2, 102.7, 72.5, $
93.1, 115.9, 83.8, 113.3, 109.4]
Max_N_Best = 3
IMSL_ALLBEST, x, y, Max_N_Best = max_n_best, /Mallows_Cp, $
Idx_Coefs = idx_coefs, $
Coefs = coefs
PRINT, '; * * * Idx_Coefs and Coefs in raw form * * *'
PRINT
PM, idx_coefs, Title = ';Idx_Coefs:'
PRINT
PM, Coefs, Title = ';Coefs'
PRINT
ntbest = max_n_best
PRINT, '; * * * How Idx_Coefs describes Coefs * * *'
PRINT
FOR i = 0, ntbest - 1 DO $
PRINT, ';regression', i+1, 'begins at row', Idx_Coefs(i),$
'; of Coefs.', FORMAT = '(a, i2, a, i2, a)'
PRINT
PRINT, ';* * * Coefs separated by ', 'regressions * * *'
PRINT
FOR i = 0, ntbest - 1 DO begin
start = idx_coefs(i)
stop = idx_coefs(i + 1) - 1
FOR j = start, stop DO begin
PRINT, coefs(j, *), FORMAT = ';(5f9.4)'
END
PRINT
END
PRINT, '; * * * Best Regressions* * *'
PRINT
FOR i = 0, ntbest - 1 DO begin
start = idx_coefs(i)
stop = idx_coefs(i + 1) - 1
count = stop - start + 1
PRINT, ';Best Regression with', count, $
';variables(s) (Mallows CP)', FORMAT = '(a, i2, a)'
PRINT, ';variable coefficient std error t p-value'
FOR j = start, stop DO $
PRINT, coefs(j, *), FORMAT = ';(i5, 2x, 4f11.4)'
PRINT
END
END
* * * Idx_Coefs and Coefs in raw form * * *
PM, Idx_Coefs
0
2
5
8
PM, Coefs
1.00000 1.46831 0.121301 12.1046 2.38419e-07
2.00000 0.662251 0.0458547 14.4424 0.00000
1.00000 1.45194 0.116998 12.4099 5.96046e-07
2.00000 0.416112 0.185611 2.24185 0.0516866
4.00000 -0.236538 0.173288 -1.36500 0.205401
1.00000 1.69589 0.204582 8.28953 1.66893e-05
2.00000 0.656915 0.0442343 14.8508 1.19209e-07
3.00000 0.250018 0.184711 1.35356 0.208889
* * * How Idx_Coefs describes Coefs * * *
regression 1 begins at row 0 of Coefs.
regression 2 begins at row 2 of Coefs.
regression 3 begins at row 5 of Coefs.
* * * Coefs separated by regressions * * *
1.0000 1.4683 0.1213 12.1046 0.0000
2.0000 0.6623 0.0459 14.4424 0.0000
1.0000 1.4519 0.1170 12.4099 0.0000
2.0000 0.4161 0.1856 2.2419 0.0517
4.0000 -0.2365 0.1733 -1.3650 0.2054
1.0000 1.6959 0.2046 8.2895 0.0000
2.0000 0.6569 0.0442 14.8508 0.0000
3.0000 0.2500 0.1847 1.3536 0.2089
* * * Best Regressions* * *
Best Regression with 2 variable(s) (Mallows CP)
variable coefficient std error t p-value
1 1.4683 0.1213 12.1046 0.0000
2 0.6623 0.0459 14.4424 0.0000
Best Regression with 3 variable(s) (Mallows CP)
variable coefficient std error t p-value
1 1.4519 0.1170 12.4099 0.0000
2 0.4161 0.1856 2.2419 0.0517
4 -0.2365 0.1733 -1.3650 0.2054
Best Regression with 3 variable(s) Mallows CP)
variable coefficient std error t p-value
1 1.6959 0.2046 8.2895 0.0000
2 0.6569 0.0442 14.8508 0.0000
3 0.2500 0.1847 1.3536 0.2089
Errors
Warning Errors
STAT_VARIABLES_DELETED: At least one variable is deleted from the full model because the variance-covariance matrix Cov is singular.
Fatal Errors
STAT_NO_VARIABLES: No variables can enter any model.
Syntax
IMSL_ALLBEST, X, Y [, ADJ_R_SQUARED=value] [, COEFS=variable] [, COV_INPUT=array] [, COV_NOBS=value] [, CRITERIONS=variable] [, /DOUBLE] [, FREQUENCIES=array] [, IDX_COEFS=variable] [, IDX_CRITERIONS=variable] [, IDX_VARS=variable] [, INDEP_VARS=variable] [, MALLOWS_CP=value] [, MAX_N_BEST=] [, MAX_N_GOOD=value] [, MAX_SUBSET=value]) [, WEIGHTS=array]
Arguments
X
Two-dimensional array containing the data for the candidate variables.
Y
One-dimensional array of length N_ELEMENTS (x(*, 0)) containing the responses for the dependent variable.
Keywords
ADJ_R_SQUARED (optional)
The adjusted R2 criterion is used, where subset sizes 1, 2, ..., N_ELEMENTS (x(*, 0)) are examined. Keywords MAX_SUBSET, ADJ_R_SQUARED, and MALLOWS_CP cannot be used together.
COEFS (optional)
Named variable into which the two-dimensional array of size (IDX_COEFS (NTBEST)) x 5 containing statistics relating to the regression coefficients of the best models is stored. Each row corresponds to a coefficient for a particular regression. The regressions are in order of increasing subset size. Within each subset size, the regressions are ordered so that the better regressions appear first. The statistic in the columns are as follows (inferences are conditional on the selected model):
- 0: variable number
- 1: coefficient estimate
- 2: estimated standard error of the estimate
- 3: t-statistic for the test that the coefficient is 0
- 4: p-value for the two-sided t test
Keywords COEFS and IDX_COEFS must be used together.
COV_INPUT (optional)
Two-dimensional square array of size (N_ELEMENTS (x(0, *)) + 1) by (N_ELEMENTS (x(0, *)) + 1) containing a variance-covariance or sum-of-squares and crossproducts matrix, in which the last column must correspond to the dependent variable.
Array COV_INPUT can be computed using IMSL_COVARIANCES. The arguments X and Y, and keywords FREQUENCIES and WEIGHTS are not accessed when this option is specified. Normally, IMSL_ALLBEST computes COV_INPUT from the input data matrices x and y. However, there may be cases when you will want to calculate the covariance matrix and manipulate it before calling IMSL_ALLBEST. See the description at the beginning of this topic for a discussion of such cases.
The keywords COV_INPUT and COV_NOBS must be used together.
COV_NOBS (optional)
Number of observations associated with array COV_INPUT. The keywords COV_INPUT and COV_NOBS must be used together.
CRITERIONS (optional)
Named variable into which the one-dimensional array of length max(IDX_CRITERIONS (NSIZE – 1), N_ELEMENTS (x(0, *)) containing in its first IDX_CRITERIONS (NSIZE – 1) elements the criterion values for each subset considered, in increasing subset size order, is stored. The keywords CRITERIONS and IDX_CRITERIONS must be used together.
DOUBLE (optional)
If present and nonzero, double precision is used.
FREQUENCIES (optional)
One-dimensional array of length N_ELEMENTS (x(*, 0)) containing the frequency for each row of x. Default: (*) = 1
IDX_COEFS (optional)
Named variable into which the one-dimensional array of length NBEST + 1 containing the locations of COEFS the first row of each of the best regressions is stored. Here, NTBEST is the total number of best regression found and is MAX_SUBSET * MAX_N_BEST if MAX_SUBSET is specified, MAX_N_BEST if either MALLOWS_CP or ADJ_R_SQUARED is specified, and MAX_N_BEST * (N_ELEMENTS (x(0, *))) otherwise. For i = 0, 1, ..., NTBEST, rows IDX_COEFS (i), IDX_COEFS(i) + 1, ..., IDX_COEFS (i + 1) – 1 of COEFS correspond to the (i + 1)-st regression. Keywords COEFS and IDX_COEFS must be used together.
IDX_CRITERIONS (optional)
Named variable into which the one-dimensional array of length NSIZE containing the locations in CRITERIONS of the first element for each subset size is stored. NSIZE is calculated as follows: NSIZE = (Max_Subset + 1) if MAX_SUBSET is set. NSIZE = (N_ELEMENTS (x(0, *)) + 1) otherwise. For i = 0, 1, ..., NSIZE – 2, element numbers IDX_CRITERIONS(i), IDX_CRITERIONS (i) + 1, ..., IDX_CRITERIONS(i + 1) – 1 of CRITERIONS correspond to the (i + 1)-st subset size. Keywords CRITERIONS and IDX_CRITERIONS must be used together.
IDX_VARS (optional)
Named variable into which the one-dimensional array of length NSIZE containing the locations in INDEP_VARS of the first element for each subset size. NSIZE is calculated as follows: NSIZE = (MAX_SUBSET + 1) if MAX_SUBSET is set. NSIZE = (N_ELEMENTS(x(0, *)) + 1) otherwise. For i = 0, 1, ..., NSIZE – 2, element numbers IDX_VARS(i), IDX_VARS (i) + 1, ..., IDX_VARS (i + 1) – 1) of INDEP_VARS correspond to the (i + 1)-st subset size. The keywords INDEP_VARS and IDX_VARS must be used together.
INDEP_VARS (optional)
Named variable into which the one-dimensional array of length IDX_VARS (NSIZE – 1) containing the variable numbers for each subset considered and in the same order as in CRITERIONS is stored. Keywords INDEP_VARS and IDX_VARS must be used together.
MALLOWS_CP (optional)
Mallows Cp criterion is used, where subset sizes 1, 2, ..., N_ELEMENTS (x(*, 0)) are examined. The keywords MAX_SUBSET, ADJ_R_SQUARED, and MALLOWS_CP cannot be used together.
MAX_N_BEST (optional)
Number of best regressions to be found. If the R2 criterion is selected, the MAX_N_BEST best regressions for each subset size examined are found. If the adjusted R2 or Mallows Cp criterion is selected, the MAX_N_BEST overall regressions are found. Default: 1
MAX_N_GOOD (optional)
Maximum number of good regressions of each subset size to be saved in finding the best regressions. Keyword MAX_N_BEST must be greater than or equal to MAX_N_BEST. Normally, MAX_N_BEST should be less than or equal to 10. It need not ever be larger than the maximum number of subsets for any subset size. Computing time required is inversely related to MAX_N_BEST. Default: 10
MAX_SUBSET (optional)
The R2 criterion is used, where subset sizes 1, 2, ..., MAX_SUBSET are examined. This option is the default with MAX_SUBSET = N_ELEMENTS (x(0, *)). Keywords MAX_SUBSET, ADJ_R_SQUARED, and MALLOWS_CP cannot be used together.
WEIGHTS (optional)
One-dimensional array of length N_ELEMENTS (x(*, 0)) containing the weight for each row of x. Default: (*) = 1
Version History