The IMSL_DISCR_ANALYSIS procedure performs a linear or a quadratic discriminant function analysis among several known groups.
This routine requires an IDL Advanced Math and Stats license. For more information, contact your sales or technical support representative.
IMSL_DISCR_ANALYSIS performs discriminant function analysis using either linear or quadratic discrimination. The output includes a measure of distance between the groups, a table summarizing the classification results, a matrix containing the posterior probabilities of group membership for each observation, and the within- sample means and covariance matrices. Linear discriminant function coefficients are also computed.
Covariance matrices are defined as follows: Let Ni denote the sum of frequencies of observations in group i and Mi denote the number of observations in group i. Then, if Si denotes the within-group i covariance matrix:
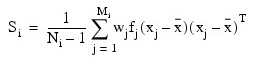
Where:
- wj is the weight of the j-th observation in group i
- fj is the frequency
- xj is the j-th observation column vector in group i
-
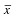 is the mean vector of the observations in group i
is the mean vector of the observations in group i
The mean vectors are computed as:
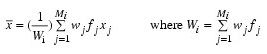
Given the means and the covariance matrices, the linear discriminant function for group i is computed as:
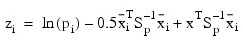
Where:
- ln (pi) is the natural log of the prior probability for the i-th group
- x is the observation to be classified
- Sp denoted the pooled covariance matrix
Let S denote either the pooled covariance matrix of one of the within-group covariance matrices Si. (S will be the pooled covariance matrix in linear discrimination, and Si otherwise.) The Mahalanobis distance between group i and group j is computed as:
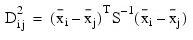
Finally, the asymptotic chi-squared test for the equality of covariance matrices is computed as follows (Morrison 1976, p. 252):
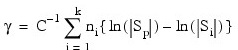
Where:
- ni is the number of degrees of freedom in the i-th sample covariance matrix
- k is the number of groups
-
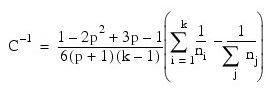
- p is the number of variables
The estimated posterior probability of each observation x belonging to group is computed using the prior probabilities and the sample mean vectors and estimated covariance matrices under a multivariate normal assumption. Under quadratic discrimination, the within-group covariance matrices are used to compute the estimated posterior probabilities. The estimated posterior probability of an observation x belonging to group i is:
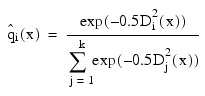
Where:
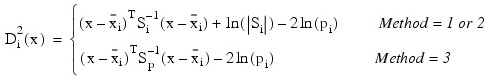
For the leaving-out-one method of classification (METHOD equal to 4, 5 or 6), the sample mean vector and sample covariance matrices in the formula for Di 2 are adjusted so as to remove the observation x from their computation. For linear discrimination (METHOD equal to 1, 2, 4, or 6), the linear discriminant function coefficients are actually used to compute the same posterior probabilities.
Using the posterior probabilities, each observation in x is classified into a group; the result is tabulated in the CLASS_TABLE array and saved in the CLASS_MEMBER array. The CLASS_TABLE array is not altered at this stage if x(i)(IDX_VARS(0)) contains a group number that is out of range. If the reclassification method is specified, then all observations with no missing values in the N_variables classification variables are classified. When the leaving-out-one method is used, observations with invalid group numbers, weights, frequencies, or classification variables are not classified. Regardless of the frequency, a 1 is added (or subtracted) from CLASS_TABLE for each row of x that is classified and contains a valid group number.
When METHOD > 3, adjustment is made to the posterior probabilities to remove the effect of the observation in the classification rule. In this adjustment, each observation is presumed to have:
- a weight of x(i)(IDX_VARS(2)) if IDX_VARS(2) > −1, and:
- a weight of 1.0 if IDX_VARS(2) = −1, and:
- a frequency of 1.0.
See Lachenbruch (1975, p. 36) for the required adjustment.
The covariance matrices are computed from their LU factorizations.
Example
The following example uses liner discrimination with equal prior probabilities on Fisher’s (1936) iris data.
.RUN
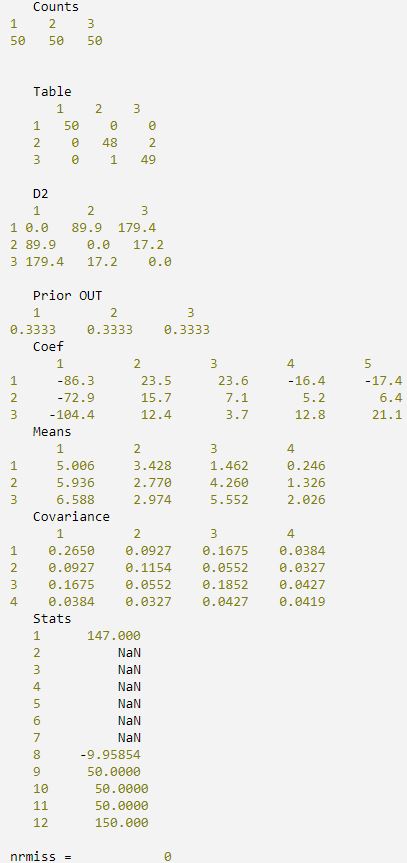
PRO print_results, counts, table, d2, prior_out, coef, means, $
cov, stats, nrmiss
num = INDGEN(3)
PRINT, ' Counts'
PRINT, num + 1, FORMAT = '(3I5)'
PRINT, counts, FORMAT = '(3I5)'
PRINT
PRINT, ' Table'
PRINT, num + 1, FORMAT = '(2X, 3I5)'
FOR i = 0, 2 DO $
PRINT, num(i) + 1, table(i, *), FORMAT = '(I2, 3I5)'
PRINT
PRINT, ' D2'
PRINT, num + 1, FORMAT = '(3I7)'
FOR i = 0, 2 DO $
PRINT, num(i) + 1, d2(i, *), FORMAT = '(I2, 3F7.1)'
PRINT
PRINT, ' Prior OUT'
PRINT, num + 1, FORMAT = '(3I10)'
PRINT, prior_out, FORMAT = '(3F10.4)'
PRINT
num = INDGEN(5)
PRINT, ' Coef'
PRINT, num + 1, FORMAT = '(1X, 5I10)'
FOR i = 0, 2 DO $
PRINT, num(i) + 1, coef(i, *), FORMAT = '(I2, 5F10.1)'
PRINT
num = INDGEN(4)
PRINT, ' Means'
PRINT, num + 1, FORMAT = '(4I10)'
FOR i = 0, 2 DO $
PRINT, num(i) + 1, means(i, *), FORMAT = '(I2, 4F10.3)'
PRINT
PRINT, ' Covariance'
PRINT, num + 1, FORMAT = '(4I10)'
FOR i = 0, 3 DO $
PRINT, num(i) + 1, cov(0, *, i), FORMAT = '(I2, 4F10.4)'
PRINT
num = INDGEN(12)
PRINT, ' Stats' FOR i = 0, 11 DO $
PRINT, num(i) + 1, stats(i)
PRINT
PRINT, 'nrmiss = ', nrmiss
END
idxv = [1, 2, 3, 4]
idxc = [0, -1, -1]
n_groups = 3
method = 3
x = IMSL_STATDATA(3)
IMSL_DISCR_ANALYSIS, x, n_groups, Idx_Vars = idxv, $
Idx_cols = idxc, Method = method, /Prior_Equal, $
Prior_Output = prior_out, Group_Counts = counts, $
Means = means, Covariances = cov, $
Coefficients = coef, Class_Member = cm, $
Class_Table = table, Prob = prob, $
Mahalanobis = d2, Stats = stats, Nmissing = nrmiss
print_results, counts, table, d2, prior_out, coef, means, $
cov, stats, nrmiss
IDL prints:
Errors
Warning Errors
STAT_BAD_OBS_1: In call #, row # of the data matrix, "x", has group number = #. The group number must be an integer between 1.0 and "n_groups" = #, inclusively. This observation will be ignored.
STAT_BAD_OBS_2: The leaving-out-one method is specified but this observation does not have a valid group number (Its group number is #). This observation (row #) is ignored.
STAT_BAD_OBS_3: The leaving-out-one method is specified but this observation does not have a valid weight or it does not have a valid frequency. This observation (row #) is ignored.
STAT_COV_SINGULAR_3: The group # covariance matrix is singular. "Stats(1)" cannot be computed. "Stats(1)" and "Stats(3)" are set to the missing value code (NaN).
Fatal Errors
STAT_COV_SINGULAR_1: The variance-covariance matrix for population number # is singular. The computations cannot continue.
STAT_COV_SINGULAR_2: The pooled variance-covariance matrix is singular. The computations cannot continue.
STAT_COV_SINGULAR_4: A variance-covariance matrix is singular. The index of the first zero element is equal to #.
Syntax
IMSL_DISCR_ANALYSIS, X, N_groups [, CLASS_MEMBER=variable] [, CLASS_TABLE=variable] [, COEFFICIENTS=variable] [, COVARIANCES=variable] [, /DOUBLE] [, GROUP_COUNTS=variable] [, IDX_COLS=array] [, IDX_VARS=array] [, MAHALANOBIS=variable] [, MEANS=variable] [, METHOD=value] [, NMISSING=variable] [, /PRIOR_EQUAL] [, PRIOR_INPUT=array] [, PRIOR_OUTPUT=variable] [, /PRIOR_PROP] [, PROB=variable] [, STATS=variable]
Arguments
N_groups (optional)
Number of groups in the data.
X
Two-dimensional array of size N_rows by N_variables + 1 containing the data, where:
- N_rows = number of rows to be processed: N_ELEMENTS(X(*,0))
- N_variables = number of variables to be used in the discrimination. The first N_variables columns correspond to the variables, and the last column contains the group numbers. The groups must be numbered 1, 2, ..., N_groups.
Keywords
CLASS_MEMBER (optional)
Named variable in the form of a one-dimensional integer array of length N_rows containing the group to which the observation was classified. If an observation has an invalid group number, frequency, or weight when the leaving-out-one method has been specified, then the observation is not classified and the corresponding elements of CLASS_MEMBER (and PROB) are set to 0.
CLASS_TABLE (optional)
Named variable in the form of a two-dimensional array of size N_groups by N_groups containing the classification table. Each observation that is classified and has a group number 1.0, 2.0, ..., N_groups is entered into the table. The rows of the table correspond to the known group membership. The columns refer to the group to which the observation was classified.
COEFFICIENTS (optional)
Named variable in the form of a two-dimensional array of size N_groups by (N_variables + 1) containing the linear discriminant coefficients. The first column of COEFFICIENTS contains the constant term, and the remaining columns contain the variable coefficients. Row i – 1 of COEFFICIENTS corresponds to group i, for i = 1, 2, ..., N_variables + 1. Array Coefficients are always computed as the linear discriminant function coefficients even when quadratic discrimination is specified.
COVARIANCES (optional)
Named variable in the form of a three-dimensional array of size g by N_variables by N_variables containing covariance results. The within-group covariance matrices (when METHOD is set to 1, 2, 4, or 5 only) is the first g-1 matrices, and the pooled covariance matrix is the g-th matrix.
DOUBLE (optional)
If present and nonzero, double precision is used.
GROUP_COUNTS (optional)
Named variable in the form of a one-dimensional integer array of length N_groups containing the number of observations in each group.
IDX_COLS (optional)
One-dimensional array containing the indices of the variables to be used in the analysis.
IDX_VARS (optional)
Three element array indicating the column numbers of X in which particular types of data are stored. Columns are numbered 0 ... N_ELEMENTS(IDX_COLS) − 1.
IDX_VARS(0) contains the index for the column of X in which the group numbers are stored.
IDX_VARS(1) and IDX_VARS(2) contain the column numbers of X in which the frequencies and weights, respectively, are stored. Set IDX_VARS(1) to a value of −1 if there will be no column for frequencies. Set IDX_VARS(2) to a value of −1 if there will be no column for weights. Weights are rounded to the nearest integer. Negative weights are not allowed.
Defaults:
- IDX_COLS = 0, 1, ..., N_variables – 1
- IDX_VARS(0) = N_variables
- IDX_VARS(1) = −1
- IDX_VARS(2) = −1
MAHALANOBIS (optional)
Named variable in the form of a two-dimensional array of size N_groups by N_groups containing the Mahalanobis distances (Dij2) between the group means. For linear discrimination, the Mahalanobis distance is computed using the pooled covariance matrix. Otherwise, the Mahalanobis distance between group means i and j is computed using the within covariance matrix for group i in place of the pooled covariance matrix.
MEANS (optional)
Named variable in the form of a two-dimensional array of size N_groups by N_variables containing the variable means. The i-th row of means contains the group i variable means.
METHOD (optional)
Method of discrimination. The method chosen determines whether linear or quadratic discrimination is used, whether the group covariance matrices are computed (the pooled covariance matrix is always computed), and whether the leaving-out-one or the reclassification method is used to classify each observation. The METHOD values are shown in the following table.
|
Method |
Discrimination method
|
Covariances computed
|
Classification method
|
|
1 (default) |
linear |
pooled, group |
reclassification
|
|
2 |
quadratic |
pooled, group |
reclassification
|
|
3 |
linear |
pooled |
reclassification
|
|
4 |
linear |
pooled, group |
leaving-out-one |
|
5 |
quadratic |
pooled, group |
leaving-out-one |
|
6 |
linear |
pooled |
leaving-out-one |
In the leaving-out-one method of classification, the posterior probabilities are adjusted so as to eliminate the effect of the observation from the sample statistics prior to its classification. In the classification method, the effect of the observation is not eliminated from the classification function.
NMISSING (optional)
Named variable into which the number of rows of data encountered containing missing values (NaN) for the classification, group, weight, and/or frequency variables is stored. If a row of data contains a missing value (NaN) for any of these variables, that row is excluded from the computations.
PRIOR_EQUAL (optional)
If present, equal prior probabilities are calculated as 1.0/N_groups. Keywords PRIOR_EQUAL, PRIOR_PROP, and PRIOR_INPUT must not be used together.
PRIOR_INPUT (optional)
If present, an array of length N_groups containing the prior probabilities for each group, such that the sum of all prior probabilities is equal to 1.0. Keywords PRIOR_EQUAL, PRIOR_PROP, and PRIOR_INPUT must not be used together.
Common choices for the Bayesian prior probabilities are given by:
- Prior_Input(i) = 1.0/N_groups (equal priors)
- Prior_Input(i) = GROUP_COUNTS/N_rows (proportional priors)
- Prior_Input(i) = Past history or subjective judgment
In all cases, the priors should sum to 1.0.
PRIOR_OUTPUT (optional)
Named variable in the form of a one-dimensional array of length N_groups containing the most recently calculated or input prior probabilities.
PRIOR_PROP (optional)
If present, prior probabilities are calculated to be proportional to the sample size in each group. Keywords PRIOR_EQUAL, PRIOR_PROP, and PRIOR_INPUT must not be used together.
PROB (optional)
Named variable in the form of a two-dimensional array of size N_rows by N_groups containing the posterior probabilities for each observation.
STATS (optional)
Named variable in the form of a one-dimensional array of length 4 + 2 * (N_groups + 1) containing various statistics of interest.
The first element is the sum of the degrees of freedom for the within-covariance matrices. The second, third, and fourth elements correspond to the chi-squared statistic, its degrees of freedom, and the probability of a greater chi-squared, respectively, of a test of the homogeneity of the within-covariance matrices (not computed if METHOD is set to 3 or 6).
The fifth through 5 + N_groups elements contain the log of the determinants of each group’s covariance matrix (not computed if METHOD is set to 3 or 6) and of the pooled covariance matrix (element 4 + N_groups). Finally, the last N_groups + 1 elements contain the sum of the weights within each group, and in the last position, the sum of the weights in all groups.
Version History