The IMSL_K_MEANS function performs a K-means (centroid) cluster analysis.
This routine requires an IDL Advanced Math and Stats license. For more information, contact your sales or technical support representative.
The IMSL_K_MEANS function is an implementation of Algorithm AS 136 by Hartigan and Wong (1979). This function computes K-means (centroid) Euclidean metric clusters for an input matrix starting with initial estimates of the K-cluster means. The IMSL_K_MEANS function allows for missing values coded as NaN (Not a Number) and for weights and frequencies.
Let p = N_ELEMENTS(x (0, *)) be the number of variables to be used in computing the Euclidean distance between observations. The idea in K-means cluster analysis is to find a clustering (or grouping) of the observations so as to minimize the total within-cluster sums-of-squares. In this case, the total sums-of-squares within each cluster is computed as the sum of the centered sum-of-squares over all non-missing values of each variable. That is:
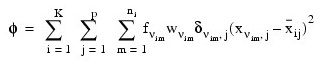
where νim denotes the row index of the m-th observation in the i-th cluster in the matrix X; ni is the number of rows of X assigned to group i; f denotes the frequency of the observation; w denotes its weight; d is 0 if the j-th variable on observation νim is missing, otherwise d is 1; and:
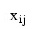
is the average of the non-missing observations for variable j in group i. This method sequentially processes each observation and reassigns it to another cluster if doing so results in a decrease of the total within-cluster sums-of-squares. See Hartigan and Wong (1979) or Hartigan (1975) for details.
Example
This example performs K-means cluster analysis on Fisher’s iris data, which is obtained by IMSL_STATDATA. The initial cluster seed for each iris type is an observation known to be in the iris type.
seeds = MAKE_ARRAY(3,4)
x = IMSL_STATDATA(3)
seeds(0, *) = x(0, 1:4)
seeds(1, *) = x(50, 1:4)
seeds(2, *) = x(100, 1:4)
cluster_group = IMSL_K_MEANS(x(*, 1:4), seeds, $
Means_Cluster = means_cluster, Ssq_Cluster= ssq_cluster, $
Counts_Cluster = counts_cluster) FORMAT = '(a, 10i4)'
FOR i = 0, 140, 10 DO BEGIN &$
PRINT, 'observation: ',i + INDGEN(10)+1, $
FORMAT = format &$
PRINT, 'cluster: ', cluster_group(i:i+9), $
FORMAT = format &$
PRINT &$
END
observation: 1 2 3 4 5 6 7 8 9 10
cluster : 1 1 1 1 1 1 1 1 1 1
observation: 11 12 13 14 15 16 17 18 19 20
cluster : 1 1 1 1 1 1 1 1 1 1
observation: 21 22 23 24 25 26 27 28 29 30
cluster : 1 1 1 1 1 1 1 1 1 1
observation: 31 32 33 34 35 36 37 38 39 40
cluster : 1 1 1 1 1 1 1 1 1 1
observation: 41 42 43 44 45 46 47 48 49 50
cluster : 1 1 1 1 1 1 1 1 1 1
observation: 51 52 53 54 55 56 57 58 59 60
cluster : 2 2 2 2 2 2 2 2 2 2
observation: 61 62 63 64 65 66 67 68 69 70
cluster : 2 2 2 2 2 2 2 2 2 2
observation: 71 72 73 74 75 76 77 78 79 80
cluster : 2 2 2 2 2 2 2 2 2 2
observation: 81 82 83 84 85 86 87 88 89 90
cluster : 2 2 2 2 2 2 2 2 2 2
observation: 91 92 93 94 95 96 97 98 99 100
cluster : 2 2 2 2 2 2 2 2 2 2
PM, [[INDGEN(3) + 1],[means_cluster]], Title = 'Cluster Means:',$
FORMAT = '(i3, 5x, 4f8.4)'
Cluster Means:
1 5.0060 3.4280 1.4620 0.2460
2 5.9016 2.7484 4.3935 1.4339
3 6.8500 3.0737 5.7421 2.0711
PM, [[INDGEN(3) + 1],[ssq_cluster]], $
Title = 'Cluster Sums of Squares:', FORMAT = '(i3, 5x, f8.4)'
Cluster Sums of Squares:
1 15.1510
2 39.8210
3 23.8795
PM, [[INDGEN(3) + 1],[counts_cluster]], Title = $
'Number of Observations per Cluster:'
Number of Observations per Cluster:
1 50
2 62
3 38
Errors
Warning Errors
STAT_NO_CONVERGENCE: Convergence did not occur.
Syntax
Result = IMSL_K_MEANS(X, Seeds [, COUNTS_CLUSTER=variable] [, /DOUBLE] [, FREQUENCIES=array] [, ITMAX=value] [, MEANS_CLUSTER=variable] [, SSQ_CLUSTER=variable] [, VAR_COLUMNS=array] [, WEIGHTS=array])
Return Value
The cluster membership for each observation is returned.
Arguments
Seeds
Two-dimensional array containing the cluster seeds, i.e., estimates for the cluster centers. The seed value for the j-th variable of the i-th seed should be in seeds (i, j).
X
Two-dimensional array containing observations to be clustered. The data value for the i-th observation of the j-th variable should be in x(i, j).
Keywords
COUNTS_CLUSTER (optional)
Named variable into which an array containing the number of observations in each cluster is stored.
DOUBLE (optional)
If present and nonzero, then double precision is used.
FREQUENCIES (optional)
One-dimensional array containing the frequency of each observation of matrix x. Default: (*) = 1
ITMAX (optional)
Maximum number of iterations. Default: 30
MEANS_CLUSTER (optional)
Named variable into which a two-dimensional array containing the cluster means is stored.
SSQ_CLUSTER (optional)
Named variable into which a one-dimensional array containing the within sum-of- squares for each cluster is stored.
VAR_COLUMNS (optional)
One-dimensional array containing the columns of x to be used in computing the metric. Columns are numbered 0, 1, 2, ..., N_ELEMENTS(x(0, *)). Default: VARS_COLUMNS(*) = 0, 1, 2, ..., N_ELEMENTS(x(0, *)) – 1
WEIGHTS (optional)
One-dimensional array containing the weight of each observation of matrix x. Default: 1
Version History
See Also
IMSL_STATDATA