The IMSL_MVAR_NORMALITY function computes Mardia’s multivariate measures of skewness and kurtosis and tests for multivariate normality.
This routine requires an IDL Advanced Math and Stats license. For more information, contact your sales or technical support representative.
The IMSL_MVAR_NORMALITY function computes Mardia’s (1970) measures b1,p and b2,p of multivariate skewness and kurtosis, respectfully, for
p = N_ELEMENTS(x(0,*)). These measures are then used in computing tests for
multivariate normality. Three test statistics, one based upon b1,p alone, one based upon b2,p alone, and an omnibus test statistic formed by combining normal scores obtained from b1,p and b2,p are computed. On the order of np3, operations are
required in computing b1,p when the method of Isogai (1983) is used, where n = N_ELEMENTS(x(*,0)). On the order of np2, operations are required in computing
b2,p.
Let:
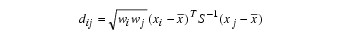
where:
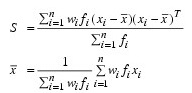
fi is the frequency of the i-th observation, and wi is the weight for this observation. (Weights wi are defined such that xi is distributed according to a multivariate normal, N(µ, Σ/wi) distribution, where Σ is the covariance matrix.) Mardia’s multivariate skewness statistic is defined as:
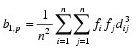
while Mardia’s kurtosis is given as:
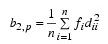
Both measures are invariant under the affine (matrix) transformation AX + D, and reduce to univariate measures when p = N_ELEMENTS(X(0,*)) = 1. Using formulas given in Mardia and Foster (1983), the approximate expected value, asymptotic standard error, and asymptotic p-value for b2,p, and the approximate expected value, an asymptotic chi-squared statistic, and p-value for the b1,p statistic are computed.
These statistics are all computed under the null hypothesis of a multivariate normal distribution. In addition, standard normal scores W1(b1,p) and W2(b2,p) (different from but similar to the asymptotic normal and chi-squared statistics above) are computed.
These scores are combined into an asymptotic chi-squared statistic with two degrees of freedom:
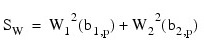
This chi-squared statistic may be used to test for multivariate normality. A p-value for the chi-squared statistic is also computed.
Example
In the following example, 150 observations from a 5 dimensional standard normal distribution are generated via routine IMSL_RANDOM. The skewness and kurtosis statistics are then computed for these observations.
m = 150
n = 5
IMSL_RANDOMOPT, set = 123457
x = FLTARR(n, m)
x(*) = IMSL_RANDOM(m*n, /Normal)
x = TRANSPOSE(x)
stats = IMSL_MVAR_NORMALITY(x, Sum_Weights = sw, Sum_Freq = sf, $
Means = means, R_Matrix = r_mat)
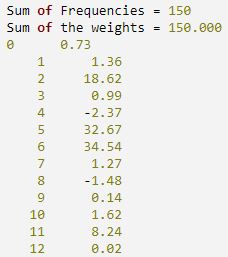
PRINT, 'Sum of Frequencies =', sf, FORMAT = '(A25, I4)'
PRINT, 'Sum of the weights =', sw, FORMAT = '(A25, F8.3)'
FOR i = 0, 12 DO PM, i, stats(i), FORMAT = '(I5, F10.2)'
IDL prints:
Syntax
Result = IMSL_MVAR_NORMALITY(X [, /DOUBLE] [, FREQUENCIES=array] [, MEANS=variable] [, NMISSING=variable] [, R_MATRIX=variable] [, SUM_FREQS=variable] [, SUM_WEIGHTS=variable] [, WEIGHTS=array])
Return Value
One-dimensional array of size 13 containing output statistics as shown in the following list:
- 0: estimated skewness
- 1: expected skewness assuming a multivariate normal distribution
- 2: asymptotic chi-squared statistic assuming a multivariate normal distribution
- 3: probability of a greater chi-squared
- 4: Mardia and Foster's standard normal score for skewness
- 5: estimated kurtosis
- 6: expected kurtosis assuming a multivariate normal distribution
- 7: asymptotic standard error of the estimated kurtosis
- 8: standard normal score obtained from Result(5) through Result(7)
- 9: p-value corresponding to Result(8)
- 10: Mardia and Foster's standard normal score for kurtosis
- 11: Mardia's SW statistic based upon Result(4) and Result(10)
- 12: p-value for Result(11)
Arguments
X
2D array containing data in which:
- N_ELEMENTS(X(*,0)) is the number of observations (numbers of rows of data) in X
- N_ELEMENTS(X(0,*)) is the dimensionality of the multivariate space for which the skewness and kurtosis are to be computed (the number of variables in X).
Keywords
DOUBLE (optional)
If present and nonzero, double precision is used.
FREQUENCIES (optional)
One-dimensional array containing the frequencies. Frequencies must be integer values. The default value is 1 for all frequencies.
MEANS (optional)
Named variable in the form of a one-dimensional array of length N_ELEMENTS(X(0,*)) containing the sample mean.
NMISSING (optional)
Named variable with the number of rows of data in X that contain missing values (NaN).
R_MATRIX (optional)
Named variable with an upper triangular array containing the Cholesky RTR factorization of the covariance matrix.
SUM_FREQS (optional)
Named variable containing the sum of the frequencies of all observations used in the computations.
SUM_WEIGHTS (optional)
Named variable containing the sum of the weights times the frequencies for all observations used in the computations.
WEIGHTS (optional)
One-dimensional array containing the weights. Weights must be non-negative. The default value is 1 for all weights.
Version History