The IMSL_RANDOM function generates pseudorandom numbers. The default distribution is a uniform (0, 1) distribution, but many different distributions can be specified through the use of keywords.
This routine requires an IDL Advanced Math and Stats license. For more information, contact your sales or technical support representative.
The IMSL_RANDOM function is designed to return random numbers from any of a number of different distributions. The determination of which distribution to generate the random numbers from is based on the presence of a keyword or groups of keywords. If IMSL_RANDOM is called without any keywords, then random numbers from a uniform (0, 1) distribution are returned.
Uniform (0,1) Distribution
The default action of IMSL_RANDOM generates pseudorandom numbers from a uniform (0, 1) distribution using a multiplicative, congruential method. The form of the generator follows:
xi ≡ cxi - 1mod (231 – 1)
Each xi is then scaled into the unit interval (0, 1). The possible values for c in the generators are 16807, 397204094, and 950706376. The selection is made by using the IMSL_RANDOMOPT procedure with the GEN_OPTION keyword. The choice of 16807 results in the fastest execution time. If no selection is made explicitly, the functions use the multiplier 16807. See IMSL_RANDOMOPT for further discussion of generator options.
The IMSL_RANDOMOPT procedure called with the Set keyword is used to initialize the seed of the random-number generator.
You can select a shuffled version of these generators. In this scheme, a table is filled with the first 128 uniform (0, 1) numbers resulting from the simple multiplicative congruential generator. Then, for each xi from the simple generator, the low-order bits of xi are used to select a random integer, j, from 1 to 128. The j-th entry in the table is then delivered as the random number, and xi, after being scaled into the unit interval, is inserted into the j-th position in the table.
The values returned are positive and less than 1.0. Some values returned may be smaller than the smallest relative spacing; however, it may be the case that some value, for example r(i), is such that 1.0 – r(i) = 1.0.
Deviates from the distribution with uniform density over the interval (a, b) can be obtained by scaling the output. See Example 3: Beta Distribution for more details.
Normal Distribution
Calling IMSL_RANDOM with keyword NORMAL generates pseudorandom numbers from a standard normal (Gaussian) distribution using an inverse CDF technique. In this method, a uniform (0,1) random deviate is generated. Then, the inverse of the normal distribution function is evaluated at that point using the IMSL_NORMALCDF function with keyword Inverse.
If the PARAMETERS keyword is specified in addition to Normal, IMSL_RANDOM generates pseudorandom numbers using an acceptance/rejection technique due to Kinderman and Ramage (1976). In this method, the normal density is represented as a mixture of densities over which a variety of acceptance/rejection methods due to Marsaglia (1964), Marsaglia and Bray (1964), and Marsaglia et al. (1964) are applied. This method is faster than the inverse CDF technique.
Deviates from the normal distribution with mean specific mean and standard deviation can be obtained by scaling the output from IMSL_RANDOM. See Example 3: Beta Distribution for more details.
Exponential Distribution
Calling IMSL_RANDOM with keyword EXPONENTIAL generates pseudorandom numbers from a standard exponential distribution. The probability density function is f(x) = e–x, for x > 0. The IMSL_RANDOM function uses an antithetic inverse CDF technique. In other words, a uniform random deviate U is generated, and the inverse of the exponential cumulative distribution function is evaluated at 1.0 – U to yield the exponential deviate.
Poisson Distribution
Calling IMSL_RANDOM with keywords POISSON and PARAMETERS = θ generates pseudorandom numbers from a Poisson distribution with positive mean θ. The probability function follows:
f(x) = (e-θθx)/x!, for x = 0, 1, 2, ...
If θ is less than 15, IMSL_RANDOM uses an inverse CDF method; otherwise, the PTPE method of Schmeiser and Kachitvichyanukul (1981) is used. (See also Schmeiser 1983.) The PTPE method uses a composition of four regions, a triangle, a parallelogram, and two negative exponentials. In each region except the triangle, acceptance/rejection is used. The execution time of the method is essentially insensitive to the mean of the Poisson.
Gamma Distribution
Calling IMSL_RANDOM with keywords GAMMA and PARAMETERS = a generates pseudorandom numbers from a Gamma distribution with shape parameter a and unit scale parameter. The probability density function follows:

Various computational algorithms are used depending on the value of the shape parameter a. For the special case of a = 0.5, squared and halved normal deviates are used; for the special case of a = 1.0, exponential deviates are generated. Otherwise, if a is less than 1.0, an acceptance-rejection method due to Ahrens, described in Ahrens and Dieter (1974), is used. If a is greater than 1.0, a 10-region rejection procedure developed by Schmeiser and Lal (1980) is used.
The Erlang distribution is a standard Gamma distribution with the shape parameter having a value equal to a positive integer; hence, IMSL_RANDOM generates pseudorandom deviates from an Erlang distribution with no modifications required.
Beta Distribution
Calling IMSL_RANDOM with keywords BETA, and PARAMETERS = [p, q] generates pseudorandom numbers from a beta distribution. With p and q both positive, the probability density function is:

where Γ(·) is the Gamma function.
The algorithm used depends on the values of p and q. Except for the trivial cases of p = 1 or q = 1, in which the inverse CDF method is used, all the methods use acceptance/rejection. If p and q are both less than 1, the method of Jöhnk (1964) is used. If either p or q is less than 1 and the other is greater than 1, the method of Atkinson (1979) is used. If both p and q are greater than 1, algorithm BB of Cheng (1978), which requires very little setup time, is used if x is less than 4, and algorithm B4PE of Schmeiser and Babu (1980) is used if x is 4 or greater. Note that for p and q both greater than 1, calling IMSL_RANDOM to generate random numbers from a beta distribution a loop getting less than four variates on each call yields the same set of deviates as executing one call and getting all deviates at once.
The values returned are less than 1.0 and greater than ε, where ε is the smallest positive number such that 1.0 – ε is less than 1.0.
Multivariate Normal Distribution
Calling IMSL_RANDOM with keywords MVAR_NORMAL and COVARIANCES generates pseudorandom numbers from a multivariate normal distribution with mean vector consisting of all zeros and variance-covariance matrix defined using keyword COVARIANCES. First, the Cholesky factor of the variance-covariance matrix is computed. Then, independent random normal deviates with mean zero and variance 1 are generated, and the matrix containing these deviates is postmultiplied by the Cholesky factor. Because the Cholesky factorization is performed in each invocation, it is best to generate as many random vectors as needed at once.
Deviates from a multivariate normal distribution with means other than zero can be generated by using IMSL_RANDOM with keywords MVAR_NORMAL and COVARIANCES, then adding the vectors of means to each row of the result.
Binomial Distribution
Calling IMSL_RANDOM with keywords BINOMIAL, PARAMETERS = [p, n] generates pseudorandom numbers from a binomial distribution with parameters n and p. Parameters n and p must be positive, and p must less than 1. The probability function (where n = Binom_n and p = Binom_p) is:

for x = 0, 1, 2, ..., n.
The algorithm used depends on the values of n and p. If n * p < 10 or p is less than machine epsilon, the inverse CDF technique is used; otherwise, the BTPE algorithm of Kachitvichyanukul and Schmeiser (see Kachitvichyanukul 1982) is used. This is an acceptance /rejection method using a composition of four regions. (TPE=Triangle, Parallelogram, Exponential, left and right.)
Cauchy Distribution
Calling IMSL_RANDOM with the keyword Cauchy generates pseudorandom numbers from a Cauchy distribution. The probability density function is:

where T is the median and T − S is the first quartile. This function first generates standard Cauchy random numbers (T = 0 and S = 1) using the technique described below, and then scales the values using T and S.
Use of the inverse CDF technique would yield a Cauchy deviate from a uniform (0, 1) deviate, u, as tan [π (u − 0.5)]. Rather than evaluating a tangent directly, however, IMSL_RANDOM generates two uniform (−1, 1) deviates, x1 and x2. These values can be thought of as sine and cosine values. If:

is less than or equal to 1, then x1/x2 is delivered as the unscaled Cauchy deviate; otherwise, x1 and x2 are rejected and two new uniform (−1, 1) deviates are generated.
This method is also equivalent to taking the ration of two independent normal deviates.
Chi-squared Distribution
Calling IMSL_RANDOM with keywords CHI_SQUARED and PARAMETERS=Df generates pseudorandom numbers from a chi-squared distribution with Df degrees of freedom.
If Df is an even integer less than 17, the chi-squared deviate r is generated as:

where n = Df /2 and the ui are independent random deviates from a uniform (0, 1) distribution. If Df is an odd integer less than 17, the chi-squared deviate is generated in the same way, except the square of a normal deviate is added to the expression above. If Df is greater than 16 or is not an integer, and if it is not too large to cause overflow in the gamma random number generator, the chi-squared deviate is generated as a special case of a gamma deviate.
Mixed Exponential Distribution
Calling IMSL_RANDOM with keywords MIX_EXPONENTIAL, and PARAMETERS = [θ1, θ2] generates pseudorandom numbers from a mixture of two exponential distributions. The probability density function is:

for x > 0.
In the case of a convex mixture, that is, the case 0 < p < 1, the mixing parameter p is interpretable as a probability; and IMSL_RANDOM with probability p generates an exponential deviate with mean θ1, and with probability 1 – p generates an exponential with mean θ2. When p is greater than 1, but less than θ1/(θ1 – θ2), then either an exponential deviate with mean θ1 or the sum of two exponentials with means θ1 and θ2 is generated. The probabilities are q = p – (p – 1) (θ1/θ2) and 1 – q, respectively, for the single exponential and the sum of the two exponentials.
Geometric Distribution
Calling IMSL_RANDOM with keywords GEOMETRIC and PARAMETERS = P generates pseudorandom numbers from a geometric distribution. The parameter P is the probability of getting a success on any trial. A geometric deviate can be interpreted as the number of trials until the first success (including the trial in which the first success is obtained). The probability function is:
f(x) = P(1 − P)x–1
for x = 1, 2, ... and 0 < P < 1.
The geometric distribution as defined above has mean 1/P.
The i-th geometric deviate is generated as the smallest integer not less than (log(Ui))/(log (1 – P)), where the Ui are independent uniform(0, 1) random numbers (see Knuth 1981).
The geometric distribution is often defined on 0, 1, 2, ..., with mean (1 – P)/P. Such deviates can be obtained by subtracting 1 from each element of the returned vector of random deviates.
Hypergeometric Distribution
Calling IMSL_RANDOM with keywords HYPERGEOMETRIC, and PARAMETERS =[M, N, L] generates pseudorandom numbers from a hypergeometric distribution with parameters N, M, and L. The hypergeometric random variable X can be thought of as the number of items of a given type in a random sample of size N that is drawn without replacement from a population of size L containing M items of this type. The probability function is:

for x = max(0, N − L + M), 1, 2, ..., min (N, M)
If the hypergeometric probability function with parameters N, M, and L evaluated at N − L + M (or at 0 if this is negative) is greater than the machine, and less than 1.0 minus the machine epsilon, then IMSL_RANDOM uses the inverse CDF technique. The routine recursively computes the hypergeometric probabilities, starting at x = max (0, N − L + M) and using the ratio:

(see Fishman 1978, p. 475).
If the hypergeometric probability function is too small or too close to 1.0, then IMSL_RANDOM generates integer deviates uniformly in the interval [1, L − i] for i = 0, 1, ..., and at the i-th step, if the generated deviate is less than or equal to the number of special items remaining in the lot, the occurrence of one special item is tallied and the number of remaining special items is decreased by one. This process continues until the sample size of the number of special items in the lot is reached, whichever comes first. This method can be much slower than the inverse CDF technique. The timing depends on N. If N is more than half of L (which in practical examples is rarely the case), You may wish to modify the problem, replacing N by L − N, and to consider the generated deviates to be the number of special items not included in the sample.
Logarithmic Distribution
Calling IMSL_RANDOM with keywords LOGARITHMIC and PARAMETERS =a generates pseudorandom numbers from a logarithmic distribution. The probability function is:

for x = 1, 2, 3, ..., and 0 < a < 1
The methods used are described by Kemp (1981) and depend on the value of a. If a is less than 0.95, Kemp’s algorithm LS, which is a “chop-down” variant of an inverse CDF technique, is used. Otherwise, Kemp’s algorithm LK, which gives special treatment to the highly probable values of 1 and 2 is used.
Lognormal Distribution
Calling IMSL_RANDOM with keywords LOGNORMAL, and PARAMETERS = [µ, σ] generates pseudorandom numbers from a lognormal distribution. The scale parameter σ in the underlying normal distribution must be positive. The method is to generate normal deviates with mean µ and standard deviation Σ and then to exponentiate the normal deviates.
The probability density function for the lognormal distribution is:

for x > 0. The mean and variance of the lognormal distribution are exp(µ + σ2/2) and exp(2µ + 2σ2) − exp(2µ + σ2), respectively.
Negative Binomial Distribution
Calling IMSL_RANDOM with keywords NEG_BINOMIAL and PARAMETERS = [r, p] generates pseudorandom numbers from a negative binomial distribution. The parameters r and p must be positive and p must be less than 1. The probability function is:

for x = 0, 1, 2, ...
If r is an integer, the distribution is often called the Pascal distribution and can be thought of as modeling the length of a sequence of Bernoulli trials until r successes are obtained, where p is the probability of getting a success on any trial. In this form, the random variable takes values r, r + 1, r + 2, ... and can be obtained from the negative binomial random variable defined above by adding r to the negative binomial variable defined by adding r to the negative binomial variable. This latter form is also equivalent to the sum of r geometric random variables defined as taking values 1, 2, 3, ...
If rp/(1 – p) is less than 100 and (1 – p)r is greater than the machine epsilon, IMSL_RANDOM uses the inverse CDF technique; otherwise, for each negative binomial deviate, IMSL_RANDOM generates a gamma (r, p/(1 – p)) deviate Y and then generates a Poisson deviate with parameter Y.
Discrete Uniform Distribution
Calling IMSL_RANDOM with keywords DISCRETE_UNIF and PARAMETERS = k generates pseudorandom numbers from a uniform discrete distribution over the integers 1, 2, ..., k. A random integer is generated by multiplying k by a uniform (0, 1) random number, adding 1.0, and truncating the result to an integer. This is equivalent to sampling with replacement from a finite population of size k.
Student’s t Distribution
Calling IMSL_RANDOM with keywords STUDENTS_T and PARAMETERS = Df generates pseudorandom numbers from a Student’s t distribution with Df degrees of freedom, using a method suggested by Kinderman et al. (1977). The method (“TMX” in the reference) involves a representation of the t density as the sum of a triangular density over (−2, 2) and the difference of this and the t density. The mixing probabilities depend on the degrees of freedom of the t distribution. If the triangular density is chosen, the variate is generated as the sum of two uniforms; otherwise, an acceptance/rejection method is used to generate the difference density.
Triangular Distribution
Calling IMSL_RANDOM with the keyword TRIANGULAR generates pseudorandom numbers from a triangular distribution over the unit interval. The probability density function is f (x) = 4x, for 0 ≤ x ≤ 0.5, and f (x) = 4 (1 – x), for 0.5 < x ≤ 1. An inverse CDF technique is used.
von Mises Distribution
Calling IMSL_RANDOM with keywords VON_MISES and PARAMETERS = c generates pseudorandom numbers from a von Mises distribution where c must be positive. The probability density function is:

for −π < x < π, where I0 (c) is the modified Bessel function of the first kind of order 0.
The probability density is equal to 0 outside the interval (−π, π).
The algorithm is an acceptance/rejection method using a wrapped Cauchy distribution as the majorizing distribution. It is due to Nest and Fisher (1979).
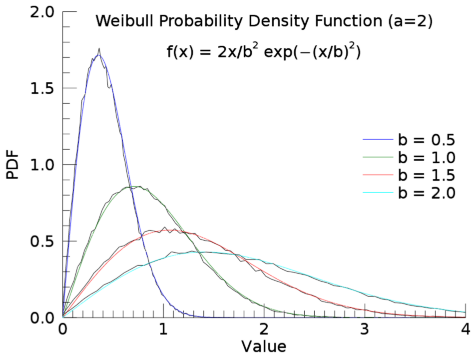
Weibull Distribution
Calling IMSL_RANDOM with keywords WEIBULL and PARAMETERS = [a, b] generates pseudorandom numbers from a Weibull distribution with shape parameter a and scale parameter b. The probability density function is:
f(x) = (a/b)(x/b)a–1exp(–(x/b)a)
for x ≥ 0, a > 0, and b > 0. The value of b is optional; if it is not specified, it is set to 1.
The IMSL_RANDOM function uses an antithetic inverse CDF technique to generate a Weibull variate; that is, a uniform random deviate U is generated and the inverse of the Weibull cumulative distribution function is evaluated at 1.0 − U to yield the Weibull deviate.
Note that the Rayleigh distribution with probability density function:
r(x) = x/σ2 exp[–x2/(2σ2)]
for x ≥ 0 is the same as a Weibull distribution with shape parameter a = 2 and scale parameter b = √2σ.
Here is a sample program to compare the random distribution with the theoretical distribution, for a=2 and b=0.5, 1.0, 1.5, and 2.0:
pro ex_weibull
defsysv,'!b', 1.0
n = 100000
ptheor = []
for i=0,3 do begin
!b = ([0.5,1.0,1.5,2])[i]
r = imsl_random(n,param=[2,!b],/weibull)
h = histogram(r,nbin=100,LOCATIONS=x)
p = plot(x,h/TOTAL(h)/(x[1]-x[0]),/overplot, axis=1, $
xtitle='Value', ytitle='PDF', font_name='Deja Vu Sans')
eqn = lambda(x:2/(!b^2)*x*exp(-(x/!b)^2))
color = (['b','g','r','c'])[i]
p1 = plot(eqn,color,/overplot,xrange=[0,4], $
name='b = ' + string(!b,'(F3.1)'))
ptheor = [ptheor, p1]
endfor
t = text(2,1.9,/DATA,'Weibull Probability Density Function (a=2)', $
align=0.5, font_name='Deja Vu Sans', clip=0)
t = text(2,1.7,/DATA,'$f(x) = 2x/b^2 exp(-(x/b)^2)$', $
align=0.5, font_name='Deja Vu Sans')
leg = legend(TARGET=ptheor, POSITION=[0.9,0.6], $
LINESTYLE='none', SHADOW=0, font_name='Deja Vu Sans')
end
Stable Distribution
Calling IMSL_RANDOM with keywords STABLE and PARAMETERS = [α, β'] generates pseudorandom numbers from a stable distribution with parameters α' and β'. α is the usual characteristic exponent parameter α, and β' is related to the usual skewness parameter β of the stable distribution. With restrictions 0 < α ≤ 2 and –1 ≤ β ≤ 1, the characteristic function of the distribution is:
ϕ(t) = exp[-|t|a exp(-π iβ (1 - |1 - α|)sign(t)/2)] for α ≠ 1
and
ϕ(t) = exp[-|t|(1 + 2iβ ln|t|)sign(t)/π)] for α = 1
When β = 0, the distribution is symmetric. In this case, if α = 2, the distribution is normal with mean 0 and variance 2; and if α = 1, the distribution is Cauchy. The parameterization using β' and the algorithm used here are due to Chambers, Mallows, and Stuck (1976). The relationship between β' and the standard β is:
β' = -tan(π (1 - α )/2) tan(-π β (1 - |1 - α|)/2) for α ≠ 1
and:
β' = β for α = 1
The algorithm involves formation of the ratio of a uniform and an exponential random variate.
Multinomial Distribution
Calling IMSL_RANDOM with keywords MULTINOMIAL, PROBABILITIES, and PARAMETERS = ntrials generates pseudorandom numbers from a K-variate multinomial distribution with parameters n and p. k = N_ELEMENTS(Probabilities) and ntrials must be positive. Each element of Probabilites must be positive and the elements must sum to 1. The probability function (with n = n, k = k, and pi = Probabilities(i)) is:


The deviate in each row of r is produced by generation of the binomial deviate x0 with parameters n and pi and then by successive generations of the conditional binomial deviates xj given x0, x1, ..., xj-2 with parameters n - x0 - x1 - ... - xj-2 and pj/(1 - p0 - p1 - ... - pj-2).
Random Points on a K-dimensional Sphere
Calling IMSL_RANDOM with the keywords SPHERE and PARAMETERS = k generates pseudorandom coordinates of points that lie on a unit circle or a unit sphere in K-dimensional space. For points on a circle (k = 2), pairs of uniform (–1, 1) points are generated and accepted only if they fall within the unit circle (the sum of their squares is less than 1), in which case they are scaled so as to lie on the circle.
For spheres in three or four dimensions, the algorithms of Marsaglia (1972) are used.
For three dimensions, two independent uniform (–1, 1) deviates U1 and U2 are generated and accepted only if the sum of their squares S1 is less than 1. Then, the coordinates:

are formed. For four dimensions, U1, U2, and S1 are produced as described above.
Similarly, U3, U4, and S2 are formed. The coordinates are then:

and:

For spheres in higher dimensions, K independent normal deviates are generated and scaled so as to lie on the unit sphere in the manner suggested by Muller (1959).
Random Permutation
Calling IMSL_RANDOM with the keyword PERMUTATION generates a pseudorandom permutation of the integers from 1 to n. It begins by filling a vector of length n with the consecutive integers 1 to n. Then, with M initially equal to n, a random index J between 1 and M (inclusive) is generated. The element of the vector with the index M and the element with index J swap places in the vector. M is then decremented by 1 and the process repeated until M = 1.
Sample Indices
Calling IMSL_RANDOM with the keywords SAMPLE_INDICES and PARAMETERS = npop generates the indices of a pseudorandom sample, without replacement, of size n numbers from a population of size npop. If n is greater than npop/2, the integers from 1 to npop are selected sequentially with a probability conditional on the number selected and the number remaining to be considered. If, when the i-th population index is considered, j items have been included in the sample, then the index i is included with probability (n - j)/(npop + 1 - i).
If n is not greater than npop/2, a O(n) algorithm due to Ahrens and Dieter (1985) is used. Of the methods discussed by Ahrens and Dieter, the one called SG* is used. It involves a preliminary selection of q indices using a geometric distribution for the distances between each index and the next one. If the preliminary sample size q is less than n, a new preliminary sample is chosen, and this is continued until a preliminary sample greater in size than n is chosen. This preliminary sample is then thinned using the same kind of sampling as described above for the case in which the sample size is greater than half of the population size. This routine does not store the preliminary sample indices, but rather restores the state of the generator used in selecting the sample initially, and then passes through once again, making the final selection as the preliminary sample indices are being generated.
Examples
Example 1
In this example, IMSL_RANDOM is used to generate five pseudorandom, uniform numbers. Since RANDOMOPT is not called, the generator used is a simple multiplicative congruential one with a multiplier of 16807.
IMSL_RANDOMOPT, Set = 123457
r = IMSL_RANDOM(5)
PM, r
The results are something like:
0.966220
0.260711
0.766262
0.569337
0.844829
Example 2
Poisson Distribution
In this example, random numbers from a Poisson distribution are computed.
IMSL_RANDOMOPT, Set = 123457
r = IMSL_RANDOM(5, /POISSON, PARAMETERS = 0.5)
PM, r
Example 3
Beta Distribution
In this example, random numbers are computed from a Beta distribution.
IMSL_RANDOMOPT, set = 123457
r = IMSL_RANDOM(5, /Beta, Parameter = [3,2])
PM, r
Example 4
Scaling the Results of IMSL_RANDOM
This example computes deviates with uniform density over the interval (10, 20) and deviates from the normal distribution with a mean of 10 and a standard deviation of 2.
IMSL_RANDOMOPT, Set = 123457
a = 10
b = 20
r = a + (b - a) * IMSL_RANDOM(5)
PM, r
The results are something like:
19.6622
12.6071
17.6626
15.6934
18.4483
stdev = 2
mean = 10
r = IMSL_RANDOM(6, /Normal) * stdev + mean
PM, r
The results are something like:
6.59363
14.4635
10.5137
12.5223
9.39352
5.71021
Example 5
Multivariate Normal Distribution
In this example, IMSL_RANDOM generates five pseudorandom normal vectors of length 2 with variance covariance matrix equal to the following:
IMSL_RANDOMOPT, SET = 123457
cov = [[.5,.375],[.375, .5]]
PM, IMSL_RANDOM(5, /MVAR_NORMAL, COVARIANCES = cov)
The results are something like:
1.45068 1.24634
0.765975 -0.0429410
0.0583781 -0.669214
0.903489 0.462826
-0.866886 -0.933426
Syntax
Result = IMSL_RANDOM(N [, /BETA] [, /BINOMIAL] [, /CAUCHY] [, COVARIANCES=value] [, /CHI_SQUARED] [, /DISCRETE_UNIF] [, /DOUBLE] [, /EXPONENTIAL] [, /GAMMA] [, /GEOMETRIC] [, /HYPERGEOMETRIC] [, /LOGARITHMIC] [, /LOGNORMAL] [, /MIX_EXPONENTIAL] [, /MULTINOMIAL] [, /MVAR_NORMAL] [, /NEG_BINOMIAL] [, /NORMAL] [, PARAMETERS=value] [, /PERMUTATION] [, /POISSON] [, PROBABILITIES=array] [, /SAMPLE_INDICES] [, /SPHERE] [, /STABLE] [, /STUDENT_T] [, /TRIANGULAR] [, /UNIFORM] [, /VON_MISES] [, /WEIBULL])
Return Value
A one-dimensional array of length n containing the random numbers. If one of the keywords SPHERE, MULTINOMIAL, or MVAR_NORMAL is used, then a two-dimensional array is returned.
Arguments
N
Number of random numbers to generate.
Keywords
BETA (optional)
If present and nonzero, the random numbers are generated from a beta distribution. Requires the PARAMETERS keyword to specify the parameters (p, q) for the distribution. The parameters p and q must be positive.
BINOMIAL (optional)
If present and nonzero, random numbers are generated from a binomial distribution. Requires PARAMETERS keyword to specify the parameters (p, n) for the distribution. The parameter n is the number of Bernoulli trials, and it must be greater than zero. The parameter p represents the probability of success on each trial, and it must be between 0.0 and 1.0.
CAUCHY (optional)
If present and nonzero, the random numbers are generated from a Cauchy distribution.
COVARIANCES (optional)
Two-dimensional, square matrix containing the variance-covariance matrix. The two-dimensional array returned by IMSL_RANDOM is of the following size:
n by N_ELEMENTS(Covariances(*, 0))
Keywords MVAR_NORMAL and COVARIANCES must be specified to return numbers from a multivariate normal distribution.
CHI_SQUARED (optional)
If present and nonzero, the random numbers are generated from a chi-squared distribution. Requires the PARAMETERS keyword to specify the parameter Df for the distribution. The parameter Df is the number of degrees of freedom for the distribution, and it must be positive.
DISCRETE_UNIF (optional)
If present and nonzero, the random numbers are generated from a discrete uniform distribution. Requires the PARAMETERS keyword to specify the parameter k for the distribution. This generates integers in the range from 1 to k (inclusive) with equal probability. The parameter k must be positive.
DOUBLE (optional)
If present and nonzero, double precision is used.
EXPONENTIAL (optional)
If present and nonzero, the random numbers are generated from a standard exponential distribution.
GAMMA (optional)
If present and nonzero, the random numbers are generated from a standard Gamma distribution. Requires the PARAMETERS keyword to specify the parameter a for the distribution. The parameter a is the shape parameter of the distribution, and it must be positive n.
GEOMETRIC (optional)
If present and nonzero, the random numbers are generated from a geometric distribution. Requires the PARAMETERS keyword to specify the parameter P for the distribution. The parameter P must be positive and less than 1.0.
HYPERGEOMETRIC (optional)
If present and nonzero, the random numbers are generated from a hypergeometric distribution. Requires the PARAMETERS keyword to specify the parameters (M, N, L) for the distribution. The parameter N represents the number of items in the sample, M is the number of special items in the population, and L is the total number of items in the population. The parameters N and M must be greater than zero, and L must be greater than both N and M.
LOGARITHMIC (optional)
If present and nonzero, the random numbers are generated from a logarithmic distribution. Requires the PARAMETERS keyword to specify the parameter a for the distribution. The parameter a must be greater than zero.
LOGNORMAL (optional)
If present and nonzero, the random numbers are generated from a lognormal distribution. Requires the PARAMETERS keyword to specify the parameters (µ, σ) for the distribution. The parameter µ is the mean of the distribution, while σ represents the standard deviation.
MIX_EXPONENTIAL (optional)
If present and nonzero, the random numbers are generated from a mixture of two exponential distributions. Requires the PARAMETERS keyword to specify the parameters (θ1, θ2, p) for the distribution. The parameters θ1 and θ2 are the means for the two distributions; both must be positive, and θ1 must be greater than θ2. The parameter p is the relative probability of the θ1 distribution, and it must be non-negative and less than or equal to θ1/( θ1 – θ2).
NEG_BINOMIAL (optional)
If present and nonzero, the random numbers are generated from a negative binomial distribution. Requires the PARAMETERS keyword to specify the parameters (r, p) for the distribution. The parameter r must be greater than zero. If r is an integer, the generated deviates can be thought of as the number of failures in a sequence of Bernoulli trials before r successes occur. The parameter p is the probability of success on each trial. It must be greater than the machine epsilon, and less than 1.0.
MULTINOMIAL (optional)
If present and nonzero, the random numbers are generated from a multinomial distribution. Requires the Parameters keyword to specify the parameter (ntrials) for the distribution, and the keyword Probabilities to specify the array containing the probabilities of the possible outcomes. The value if ntrials is the multinomial parameter indicating the number of independent trials.
MVAR_NORMAL (optional)
If present and nonzero, the random numbers are generated from a multivariate normal distribution. Keywords MVAR_NORMAL and COVARIANCES must be specified to return numbers from a multivariate normal distribution.
NORMAL (optional)
If present and nonzero, the random numbers are generated from a standard normal distribution using an inverse CDF method.
PARAMETERS (optional)
Specifies parameters for the distribution used by IMSL_RANDOM to generate numbers. Some distributions require this keyword to execute successfully. The type and range of these parameters depends upon which distribution is specified. See the keyword for the desired distribution or the description at beginning for more details.
Note: The keywords A, Pin, Qin, and Theta are still supported, but are now deprecated. Please use the PARAMETERS keyword instead.
PERMUTATION (optional)
If present and nonzero, then generate a pseudorandom permutation.
POISSON (optional)
If present and nonzero, the random numbers are generated from a Poisson distribution. Requires the PARAMETERS keyword to specify the parameter θ for the distribution. The parameter θ represents the mean of the distribution, and it must be positive.
PROBABILITIES (optional)
Specifies the array containing the probabilities of the possible outcomes. The elements of P must be positive and must sum to 1.0.
Keywords MULTINOMIAL and PROBABILITIES must be specified to return numbers from a Multinomial distribution.
SAMPLE_INDICES (optional)
If present and nonzero, generate a simple pseudorandom sample of indices. Requires the PARAMETERS keyword to specify the parameter npop, the number of items in the population.
SPHERE (optional)
If present and nonzero, the random numbers are generated on a unit circle or K-dimensional sphere. Requires the PARAMETERS keyword to specify the parameter k, the dimension of the circle (k = 2) or of the sphere.
STABLE (optional)
If present and nonzero, the random numbers are generated from a stable distribution. Requires the PARAMETERS keyword to specify the parameters A and bprime for the stable distribution. A is the characteristic exponent of the stable distribution. A must be positive and less than or equal to 2. bprime is related to the usual skewness parameter b of the stable distribution.
STUDENT_T (optional)
If present and nonzero, the random numbers are generated from a Student’s t distribution. Requires the PARAMETERS keyword to specify the parameter Df for the distribution. The Df parameter is the number of degrees of freedom for the distribution, and it must be positive.
TRIANGULAR (optional)
If present and nonzero, the random numbers are generated from a triangular distribution.
UNIFORM (optional)
If present and nonzero, the random numbers are generated from a uniform (0, 1) distribution. The default action of this returns random numbers from a uniform (0, 1) distribution.
VON_MISES (optional)
If present and nonzero, the random numbers are generated from a von Mises distribution. Requires the PARAMETERS keyword to specify the parameter c for the function. The parameter c must be greater than one-half the machine epsilon.
WEIBULL (optional)
If present and nonzero, the random numbers are generated from a Weibull distribution. Requires the PARAMETERS keyword to specify the parameters (a, b) for the distribution. The parameter a is the shape parameter, and it is required. The parameter b is the scale parameter, and is optional (Default: b = 1.0).
Version History
See Also
Overview of Random Number Generation